Human pathogens, such as bacteria, use multiple strategies to adhere to a host cell. Both Gram-positive and Gram-negative bacteria use specialized protein filaments that can anchor to the cell membrane. These extracellular hair-like appendages, known as bacterial pili or fimbriae, are subjected to a mechanical stress reaching several hundred picoNewtons caused by the continual fluid flow and attempt to detach or tear the bacterial adhesion. For example, bacteria such as E. coli is able to withstand fluid forces in the bladder, otherwise it would be rapidly removed during urination. Thus, pilus may have adhesion strategies and specific mechanical properties that are adapted to the rheological properties of the media in which these bacteria function.
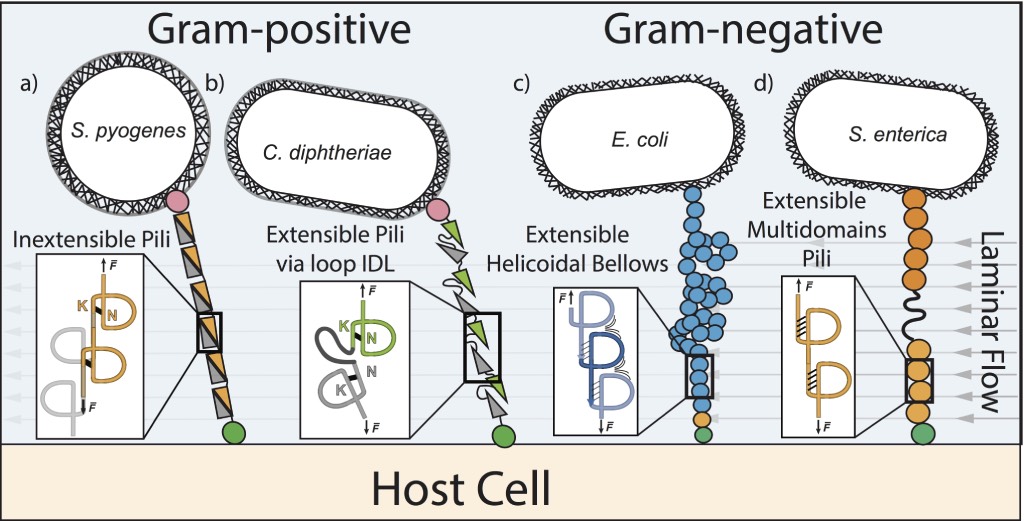
In Gram-negative bacteria, such as E. coli, the pilus proteins are exclusively concatenated by hydrogen bonds. However, in Gram-positive bacteria, such as S. pyogenes, the proteins are covalently attached to one another through the action of a sortase, which catalyzes the polymerization reaction. Thus, Gram-positive pili are inherently considerably stronger than Gram-negative pili. This is an initial major variation that allows bacterial pilus to be classified based on the organization of their structural pili. Additionally, a novel evolutionary characteristic occurs in the structural protein of the pilus in Gram-positive bacteria; an intramolecular bond is present between the lysine’s side chain and asparagine residue. This generates the formation of an isopeptide bond, preventing mechanical protein unfolding. However, subtle differences occur between Gram-positive pilus. In some cases the isopeptide bond is between the first and last beta strand of the pili protein domain (Fig. a) while in other cases the bacteria generate a more complex arrangement; two adjacent domains, where each domain has an internal isopeptide bond, shares an unstructured loop that stretches when subjected to mechanical stress (Fig. b). Thus, two types of pilus can be distinguished in Gram-positive bacteria: inextensible and extensible pili. The respective consequences of the bacterial adhesion between inextensible and extensible pili lack both deeper investigation and knowledge.
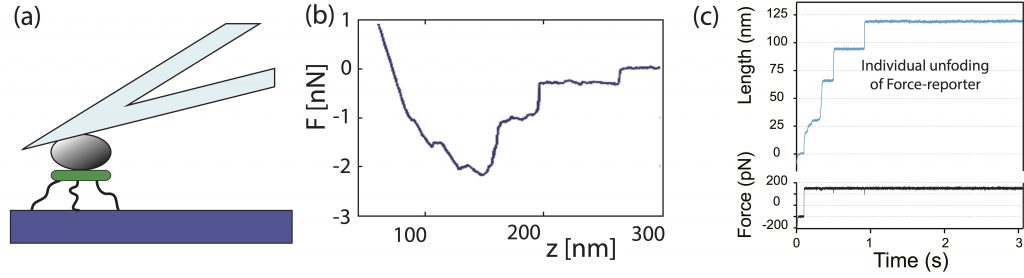
In SMAT-C we have available several techniques for the single molecules testing, magnetic tweezers, optical tweezer, and atomic force spectroscopy. See below.
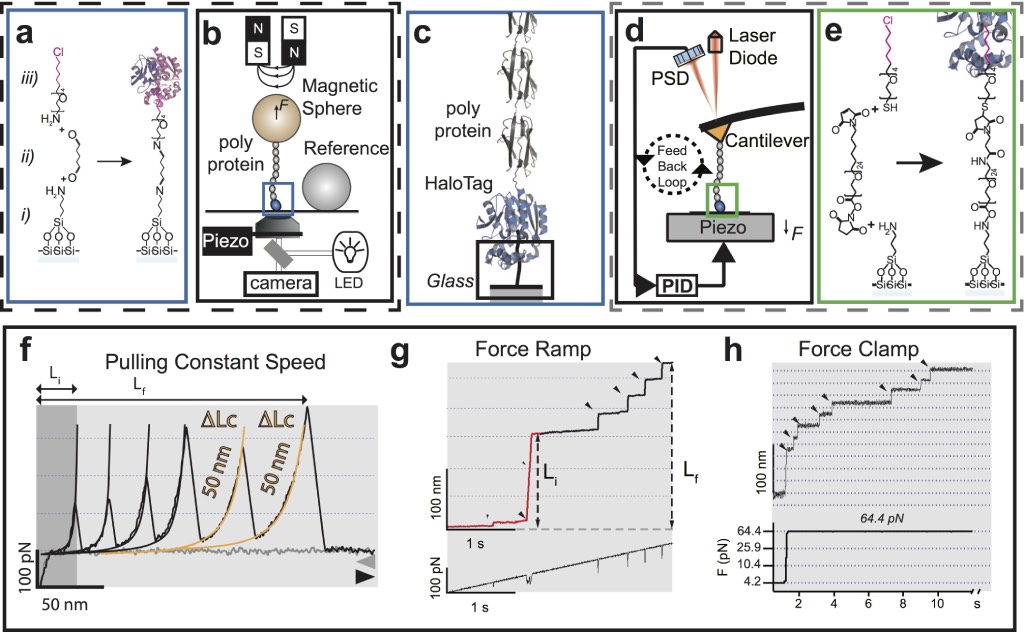
Magnetic Tweezers and AFM spectrometers for the manipulation and application of force to individual molecules. Both the magnetic clamps (a-c) and the atomic force microscope (c-e) use the HaloTag methodology for the covalent anchorage of molecules Popa_2013. The HaloTag chemistry consists of only three steps: silanization (i); addition of an amino-glutaraldehyde crosslinking agent or NHS-Maleimide-PEG- (ii); and finally, the addition of the enzyme HaloTag ligand, the chloro-alkane. b) the MT consists of three main components: magnets positioner (voice-coil), a piezo-objective, and a video camera Popa_2016, while the AFM (d) requires the implementation of an analog feedback system for ramp of force and constant force protocols (see g and h). c) in both instruments, MT and AFM, the designed HaloTag fusion polyprotein recognizes the immobilized alkane on the glass, generating a covalent immobilization to the glass. f-h) results recorded under the three modalities of mechanical disturbance. f) constant speed protocol (FX), where each peak indicates domain unfolding within a polyprotein. g) force ramp (F-ramp) that indicates both the force and the unfolding kinetics. Here, unfolding is identified as an extension increase within the results. h) constant force protocol (FC), which facilitates the effective determination of the polyprotein’s unfolding speed constant. As before, domain unfolding is identified within the results as an extension increase.